P83: Development of peptide-antibiotic conjugate therapeutics for improved targeting and killing of bacteria
Project abstract
The goal of this project is to develop new antibacterial conjugate therapeutics that combine potent small-molecule antibiotics with recently developed antimicrobial peptides (AMPs) that target bacteria with high selectivity.
The low cost and broad global availability of antibiotics has reduced bacterial infections to one of the least common causes of death today. However, due to systematic overuse in humans, livestock, and crops current antibiotics are failing rapidly [1]. At the same time only two new classes of antibiotics have been developed for clinical use since the 1970s, and all current antibiotics have side effects stemming from their indiscriminate killing of large numbers of symbiotic bacteria. Hence there is a huge unmet need to develop a new generation of antimicrobials that evade resistance formation and target pathogenic bacterial species specifically without harming the human tissues or the microbiome.
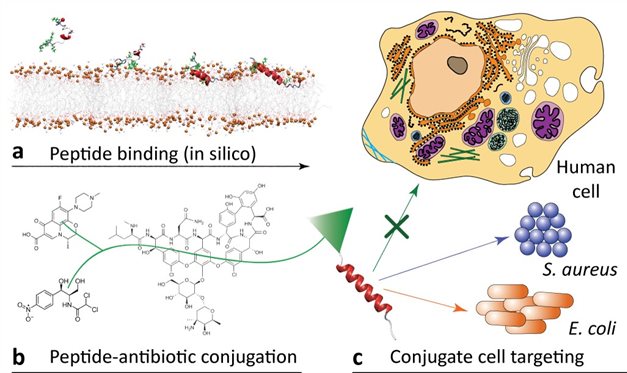
We have recently developed a simulation-guided design methodology that allows the design of short AMPs that bind selectively to specific pathogenic bacteria without binding to human cells [2]. Here we propose to apply this approach to develop AMPs with highly selective binding towards specific pathogenic bacteria and to conjugate these new AMPs to approved small-molecule antibiotics. Combining the precise targeting capability of the AMPs with the killing-power of small-molecule antibiotics has the potential to (i.) improve antibiotic potency by delivering the drug directly to the target pathogen, (ii.) reduce side-effects by selectively targeting only pathogenic bacteria, and (iii.) allow increased dosing to overcome drug-resistance by avoiding toxicity due to distribution of the conjugates in healthy tissues.
Aims
While many drug-resistant strains can still be killed by antibiotics, the doses required are typically toxic, as well as damaging to the human microbiome. In our preliminary work we have shown that molecular dynamics simulations can reveal the mechanisms of antimicrobial activity [3] and can be used to guide the design of new AMPs that bind strongly to specific bacterial strains with high selectivity [1]. Here we propose to improve the precision of bacterial targeting of these AMPs and conjugate them with small-molecule antibiotics. This has the potential to greatly reduce side effects by killing only target strains, and improve antimicrobial efficiency by allowing much higher dosing, which will allow targeting drug-resistant strains. The overall goal of the project is to generate the data necessary for translation of our compounds into the clinic. The proposed work is split into four aims:
- Training in computational techniques. You will learn molecular modelling and simulation techniques to set up, run, and analyse atomic (Ulmschneider lab) and coarse-grain molecular dynamics simulations (Lorenz lab). Also you will be trained in various rare-event sampling methods that will allow us to understand the free energy landscapes of the interactions between the conjugated peptides and bacterial membranes (Ulmschneider lab). The simulation work will be complimented by ongoing experimental work in which the peptides will be synthesized and conjugated them with small molecule antibiotics, and then investigated with a battery of state-of-the-art experimental biophysical techniques (e.g. fluorescence spectroscopy and microscopy) and cell culture assays to characterise these compounds (Ulmschneider and Cobb labs).
- Simulation-guided design of antimicrobial peptide templates with good bacterial binding signatures. You will construct bacterial membrane models and set up partitioning simulations to guide the design of peptides with improved selectivity for specific bacterial membranes in an iterative approach. These template designs will be further improved by screening of synthetic peptide libraries against bacterial membranes, which will utilise various rare-event sampling methodologies to investigate the mechanism by which the antimicrobial peptides attack the model membranes.
- Conjugation of bacteria-selective AMPs with small molecule antibiotics. Potent bacteria-selective AMPs will be conjugated to approved small-molecule antibiotics (vancomycin, levofloxacin, chloramphenicol and neomycin B). Theser antibiotics have previously been shown to be active when attached to various polypeptides, using a covalent chemical linking strategies (amide and CuAAC).
The simulation work carried out in this project will also be compared to an experimental evaluation of antibacterial activity and human cell toxicity of the peptide-drug conjugate. Concentration-dependant antibacterial activity of the AMP-antibiotic conjugates will be tested using an in vitro growth inhibition and sterilisation assays on E.coli, S.aureus, P.aeroginosa and methillicin resistant S.aureus (MRSA). AMP-antibiotic conjugate toxicity will be tested using human cell lines and compared to BTP-only and antibiotic-only antibacterial activities and toxicities. Therefore the students will be able to learn about these various experimental methods and see the full method and see the full process of transferring knowledge from simulations to molecular-scale and finally cell-level experimental testing.
Reference 1: Zaman, S. B. et al. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 9, (2017).
Reference 2: International patent filed on 12th March 2018 (application #: PCT/US18/21985): Chen & Ulmschneider, “Antimicrobial peptides and methods of making and using same”.
Reference 3: Y. Wang, C.H. Chen, D. Hu, M.B. Ulmschneider, & J.P. Ulmschneider. Spontaneous formation of structurally diverse membrane channel architectures from a single antimicrobial peptide. Nature Communications 2016, 7, (13535), 1-9
Reference 4: J.P. Ulmschneider & M.B. Ulmschneider. Molecular dynamics simulations are redefining our view of peptides interacting with biological membranes. Account of Chemical Research 2018, 51 (5), 1106-1116
