I compare accumulation of these proteins to global warming, where we’re all contributing a tiny bit too much to greenhouse gas emissions and that adds up to cause a huge problem.
Professor Jernej Ule, senior author on the paper
24 September 2025
Interstasis: how cells keep groups of proteins in check to prevent toxicity
New research from the Francis Crick Institute and UK Dementia Research Institute (UKDRI) at King’s College London has discovered a new mechanism for gene expression control that prevents toxic protein accumulation in cells.

The study, published in Nature, identified ‘interstasis’: a mechanism by which proteins regulate their own gene expression. This research crucially shines a light on how cells prevent toxic levels of protein clustering and opens directions for future research into how this regulation may go wrong in diseases such as Alzheimer’s.
The process of gene expression controls the levels of proteins in all the cells in our body. Cells must carefully control the levels of these proteins. Failing to do so can be toxic for the cell and has been linked to age-related neurodegenerative diseases like Alzheimer’s Disease. However, for some genes over-expression is more harmful than for others. Researchers wanted to explore why certain genes are harmful when they are overexpressed. And how the cell controls the levels of the proteins made by these genes.
The research addressed these questions by looking at genes that have been previously linked to toxic over-expression. These genes encode condensation-prone proteins, so named because when protein expression reaches certain levels, they cluster into a semi-solid known as a condensate.
The genes that encode proteins found in condensates have a key shared feature – they contain large unstructured regions. While over-expression of these genes is linked to toxicity, normally cellular levels of condensation-prone proteins are tightly regulated, preventing toxicity.
Researchers sought to explore how cells normally regulate levels of condensation-prone proteins. The findings unveiled a previously unknown cellular regulatory mechanism called ‘interstasis’ by which proteins regulate their own gene expression, as well as the expression of other genes whose proteins also form part of the condensate. These proteins sense the level of condensation-prone protein expression by detecting levels of arginine, a charged component of these proteins. Once arginine levels reach a certain level, the condensation-prone proteins trap their own mRNAs (the mid-stage of gene expression) into speckles (a form of condensate), preventing the mRNAs from leaving the nucleus and being translated into more condensation-prone proteins. “When the amounts of these proteins in speckles were increased, we saw a build-up of their own mRNAs also within the speckles. And this decreased further production of these proteins, thus promoting their collective equilibrium,” explains Dr Neve Costello Heaven, one of the lead authors on the project.
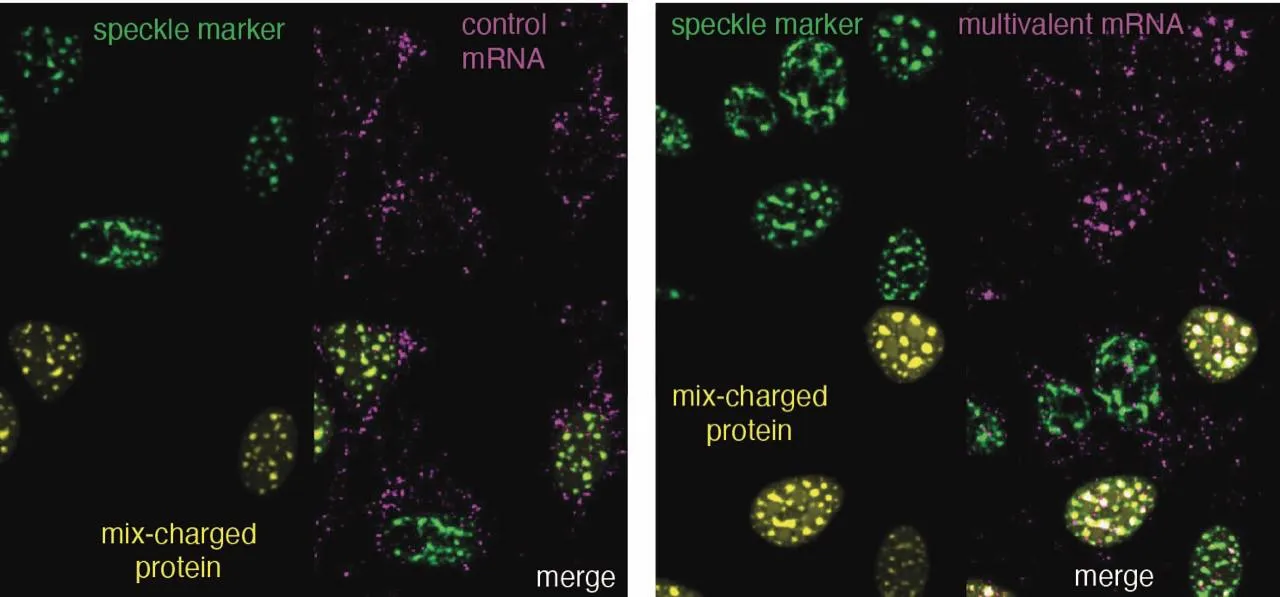
But how do the condensation-prone proteins attract their own mRNAs? The team discovered that this selectivity is due to a bias towards certain genetic codons (‘three-letter codes’) that promote repetitive sequences in their own mRNAs. These repetitive regions bind specific RNA binding proteins, which help to engage the regulatory mechanism of interstasis.
“This project was fascinating because it shows how evolution can exploit a very basic and fundamental property of the genetic code to create an elegant regulatory mechanism which keeps the protein levels in the cell in balance,” explains Dr Rupert Faraway, a first author on the paper.
Future research will focus on the role interstasis plays in aging and neurodegeneration, as well as possible therapeutic targets.
What’s really interesting is that this balance is often disrupted during the aging process and in a variety of diseases, including Alzheimer's disease, so we're now looking to investigate how this balancing act fails in those situations and how it might be restored.
Dr Rupert Faraway, co-first author on the paper
Collective homeostasis of condensation-prone proteins via their mRNAs (Faraway, Costello Heaven et al.) (DOI 10.1038/s41586-025-09568-w) was published in Nature.
This research is also reported on the Francis Crick Institute Website
This research was performed at the Francis Crick Institute and funded by the European Research Council, the Wellcome Trust, the UK Dementia Research Institute and the Francis Crick Institute (which receives funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust).
This research was conducted with support from the Scientific Computing, Advanced Sequencing, Genomics, Advanced Light Microscopy, Proteomics and Cell Sciences team at the Crick, and the National Institute of Chemistry in Ljubljana, Slovenia.
For more information please contact Patrick O’Brien (Media Manager)

