
Dr Alessandra Vigilante
Reader in Bioinformatics
- Head of the Hub for Applied Bioinformatics (HAB)
- Course Director MSc Applied Bioinformatics
Research interests
- Cell Biology
- Medicine
Biography
Alessandra Vigilante (AV) is a Reader in Bioinformatics at the Center for Gene Therapy and Regenerative Medicine with a focus on genotype- phenotype interactions and data integration. AV obtained her PhD in Bioinformatics in Naples (2008-2011) before moving the UK to join Nicholas Luscombe group first at the EMBL-European Bioinformatics Institute as a visiting student (2011-2012) and then as a postdoctoral fellow at UCL (2012-2017). Alessandra Vigilante’s group has significant expertise and experience in the analysis and integration of large scale genomic, epigenomic and transcriptomic data (i.e. single-cell RNA-seq and ATAC-seq datasets, ChIP-seq etc…), and in the implementation of novel computational methods for various bespoke analyses to gain biological insights. AV is actively involved in a great network of collaborations to develop multidisciplinary approaches to research efforts, working with faculty members within King’s and other research institutes.
Alessandra is the Head of Hub for Applied Bioinformatics (HAB), a new Faculty initiative aiming to enhance bioinformatics networking, support key research projects and provide training and engagement activities.
Alessandra teaches biomedical undergraduate students and established the interdisciplinary MSc in Applied Bioinformatics. Committed to supporting early career researchers, Alessandra has successfully supervised and mentored many BSc, MSc, and PhD students in bioinformatics.
Research

Vigilante Lab
Our primary research areas can be broadly classified as: Use of integration approaches to uncover genotype–phenotype interactions; and,Use of single-cell RNA sequencing to study developmental and stem cell biology
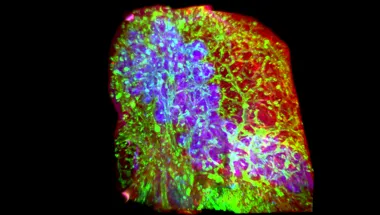
From Developmental Biology to Regenerative Medicine
Understanding organ development and tissue regeneration provides a framework for elucidating disease mechanisms as well as for developing new therapeutics.

The Hub for Applied Bioinformatics (HAB)
Welcome to the Hub for Applied Bioinformatics @KCL, a place where big data are transformed into valuable knowledge that can drive scientific breakthroughs in the fields of life sciences and medicine.
News
King's College London Awarded £2.6M MRC Programme Grant to Advance Research into Food Allergy and Oral Tolerance in Early Life
King’s College London researchers have been awarded a prestigious £2.6 million Programme Grant from the Medical Research Council (MRC) Infections and Immunity...

King's set to launch new master's course on Applied Bioinformatics
The Faculty of Life Sciences & Medicine will be launching the new postgraduate MSc course for 2024 entry, aiming to provide the skills to analyse, interpret...
Features
5 minutes with Alessandra Vigilante
Dr Alessandra Vigilante is a Senior Lecturer in Bioinformatics in the Centre for Gene Therapy and Regenerative Medicine within our School of Basic & Medical...

Research

Vigilante Lab
Our primary research areas can be broadly classified as: Use of integration approaches to uncover genotype–phenotype interactions; and,Use of single-cell RNA sequencing to study developmental and stem cell biology

From Developmental Biology to Regenerative Medicine
Understanding organ development and tissue regeneration provides a framework for elucidating disease mechanisms as well as for developing new therapeutics.

The Hub for Applied Bioinformatics (HAB)
Welcome to the Hub for Applied Bioinformatics @KCL, a place where big data are transformed into valuable knowledge that can drive scientific breakthroughs in the fields of life sciences and medicine.
News
King's College London Awarded £2.6M MRC Programme Grant to Advance Research into Food Allergy and Oral Tolerance in Early Life
King’s College London researchers have been awarded a prestigious £2.6 million Programme Grant from the Medical Research Council (MRC) Infections and Immunity...

King's set to launch new master's course on Applied Bioinformatics
The Faculty of Life Sciences & Medicine will be launching the new postgraduate MSc course for 2024 entry, aiming to provide the skills to analyse, interpret...

Features
5 minutes with Alessandra Vigilante
Dr Alessandra Vigilante is a Senior Lecturer in Bioinformatics in the Centre for Gene Therapy and Regenerative Medicine within our School of Basic & Medical...

