Martin Ulmschneider’s group studies how peptides and proteins interact with cellular membranes and carry out their biological functions. Despite being essential for a wide range of critical cellular processes the functional mechanics of many membrane active peptides and proteins remain poorly understood. The group uses both computational and experimental techniques to reveal the molecular mechanisms and atomic detail interactions driving membrane active peptide and protein function in biological lipid bilayers, as well as to design and optimize synthetic membrane active peptides for biomedical applications.
Projects
Development of new molecular simulation algorithms to map drug delivery into tissues
Efficient delivery of drugs into target tissues is essential for therapeutic success. At present, the relationship between drug chemistry and the spatial distribution of drugs in tissues is poorly understood, chiefly due to considerable challenges associated with imaging drugs at high spatio-temporal resolution in live tissues. Our goal is to build on advances in molecular simulation technology to capture the process of drug diffusion from the vasculature into surrounding tissues with sub-micrometer resolution. The goal is to construct a framework for precise calibration of drug dosing regimens, allowing optimization of drug chemistries for target tissue uptake.
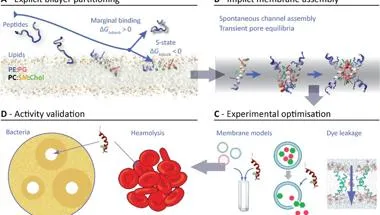
Developing a design platform for developing antimicrobial peptides
Developing new drugs to tackle rapidly rising antibiotic resistance is one of the most pressing and critical unmet needs in global healthcare. Antimicrobial peptide (AMPs) are extremely promising pharmacophores, but their chemical diversity, flexible nature, and prohibitive number of possible configurations, combined with the lack of suitable design and optimization tools has impeded translation for clinical use. This project, summarised in the schematic, seeks to develop and experimentally validate quantitatively accurate in silico tools to accelerate rational design, optimization, and characterisation of promising new AMPs. In particular, we focus on a combination of genetic and artificial intelligence algorithms, mechanistic simulations, and experimental screening tools to enable rapid tuning of AMPs to target specific species in the microbiome. The ultimate goal is to develop a set of computational tools to rapidly and accurately identify strong leads from very large candidate libraries for downstream pre-clinical evaluation.
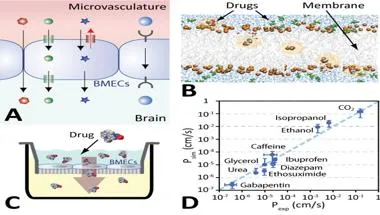
Developing technology for rapidly predicting brain exposure of drugs
Treating central nervous system (CNS) disorders is complicated by the blood-brain barrier (A), which limits CNS uptake of most chemistries. This is impeding drug development efforts as in vitro and in vivo brain exposure assays are costly and time-consuming. We are working on new experimentally validated (C) in silico methods to rapidly predict brain exposure of drugs quantitatively (D). The goal is to develop a reliable in silico screening tool that provides unprecedented insights into the transport mechanisms (B) and enables pre-synthesis lead optimization of drugs for CNS delivery.
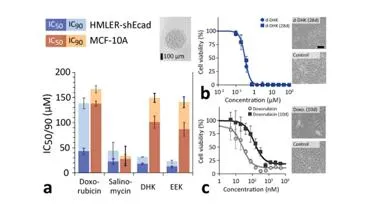
Development of nano-formulated peptide chemotherapies
Cancer is the leading cause of death in the UK. Despite considerable reduction in mortality rates over the last 30 years, long-term prognosis remains poor especially for aggressive and late-stage cancers, which have limited treatment options. Our goal is to develop new in silico and in vitro tools to bring the promising class of membrane-active peptides into the arms race against cancer. Our first-generation compounds (EEK) show promising activity and reduced toxicity compared to the standard of care (doxorubicin) (a). In addition, this class of compounds does not seem to induce drug resistance (b), a common problem with current chemotherapeutics (c).
Projects

Development of new molecular simulation algorithms to map drug delivery into tissues
Efficient delivery of drugs into target tissues is essential for therapeutic success. At present, the relationship between drug chemistry and the spatial distribution of drugs in tissues is poorly understood, chiefly due to considerable challenges associated with imaging drugs at high spatio-temporal resolution in live tissues. Our goal is to build on advances in molecular simulation technology to capture the process of drug diffusion from the vasculature into surrounding tissues with sub-micrometer resolution. The goal is to construct a framework for precise calibration of drug dosing regimens, allowing optimization of drug chemistries for target tissue uptake.

Developing a design platform for developing antimicrobial peptides
Developing new drugs to tackle rapidly rising antibiotic resistance is one of the most pressing and critical unmet needs in global healthcare. Antimicrobial peptide (AMPs) are extremely promising pharmacophores, but their chemical diversity, flexible nature, and prohibitive number of possible configurations, combined with the lack of suitable design and optimization tools has impeded translation for clinical use. This project, summarised in the schematic, seeks to develop and experimentally validate quantitatively accurate in silico tools to accelerate rational design, optimization, and characterisation of promising new AMPs. In particular, we focus on a combination of genetic and artificial intelligence algorithms, mechanistic simulations, and experimental screening tools to enable rapid tuning of AMPs to target specific species in the microbiome. The ultimate goal is to develop a set of computational tools to rapidly and accurately identify strong leads from very large candidate libraries for downstream pre-clinical evaluation.

Developing technology for rapidly predicting brain exposure of drugs
Treating central nervous system (CNS) disorders is complicated by the blood-brain barrier (A), which limits CNS uptake of most chemistries. This is impeding drug development efforts as in vitro and in vivo brain exposure assays are costly and time-consuming. We are working on new experimentally validated (C) in silico methods to rapidly predict brain exposure of drugs quantitatively (D). The goal is to develop a reliable in silico screening tool that provides unprecedented insights into the transport mechanisms (B) and enables pre-synthesis lead optimization of drugs for CNS delivery.

Development of nano-formulated peptide chemotherapies
Cancer is the leading cause of death in the UK. Despite considerable reduction in mortality rates over the last 30 years, long-term prognosis remains poor especially for aggressive and late-stage cancers, which have limited treatment options. Our goal is to develop new in silico and in vitro tools to bring the promising class of membrane-active peptides into the arms race against cancer. Our first-generation compounds (EEK) show promising activity and reduced toxicity compared to the standard of care (doxorubicin) (a). In addition, this class of compounds does not seem to induce drug resistance (b), a common problem with current chemotherapeutics (c).






